Betafab GUI¶
The graphical user interface of the β-peptide builder can be found in the Plugins menu of PyMOL. The main window is shown in the figure below.
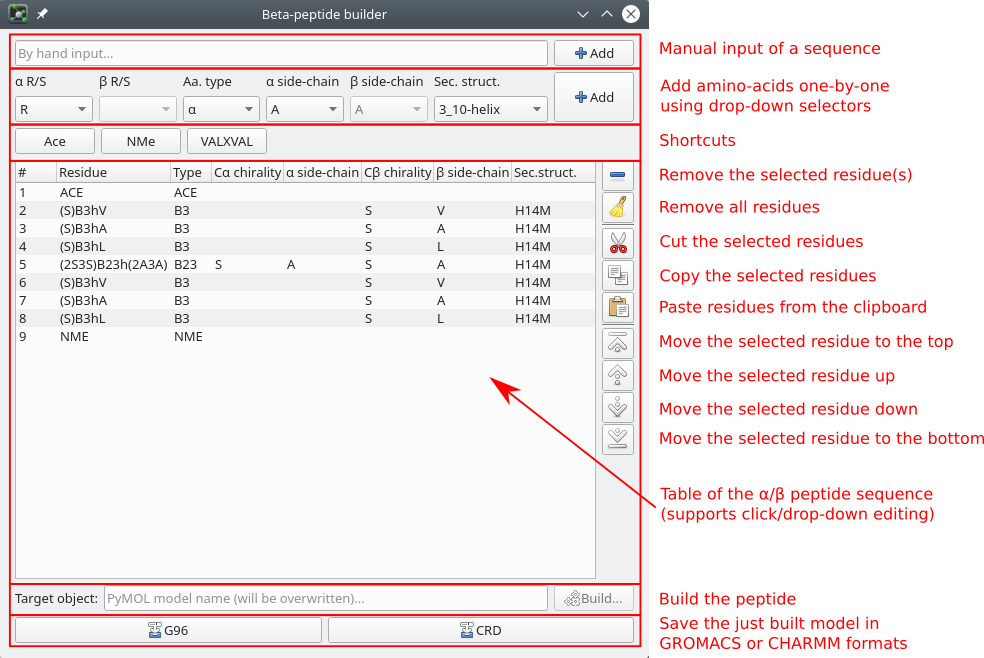
The Betafab GUI window explained
Building blocks (α- or β-amino acids, as well as Ace and NMe capping groups) can be added in three different ways to the sequence (top of the window):
- manual input of either a simple amino-acid or a whole sequence (comma separated amino acid abbreviations). The notation is the same as used by the command betafab2, see Simplified nomenclature
- one-by-one addition of amino acids using the drop-down selectors.
- using the quick-access buttons (currently for Ace and NMe capping groups and a test peptide)
The central part shows the peptide sequence and allows a simple means for editing. Amino acids can be removed, reordered and even modified. By double-clicking on cells in columns 3-8, properties of the residue (type, sidechains, stereoisomers) can be changed using drop-down selectors.
The desired secondary structure can also be given in the last column. The drop-downs display the list of the known secondary structures in the Secondary structure database.
The peptide sequence can be built by supplying a PyMOL model name for it and pressing the “Build” button.
The built model can be saved to .g96 or .crd files using the corresponding push buttons.